|
---------------------------- 2017 Platform Presentation Winner --------------------------
[1278] Ovarian Serous Carcinomas with Mixed Features of High Grade and Low Grade Serous Carcinoma Display Heterogeneous Genome Wide Copy Number Variation Profiles
Shabnam Zarei, Chen Wang, Jean-Pierre A Kocher, Yan Wang, Debra A Bell, Sarah E Kerr.
Background: Classic high and low grade serous carcinomas (HGSC and LGSC) are thought to be distinct at a molecular level. HGSC have TP53 mutations (96%) and widespread copy number changes, whereas LGSC have KRAS, BRAF and PTEN mutations and are near diploid. Little is known about the molecular profi le when mixed morphologic features of HGSC and LGSC are seen in the same patient. We previously showed that majority of these cases (70%) do not show TP53, KRAS, BRAF or PTEN mutations. In this study we describe genome wide copy number variation (CNV) in such tumors.
Design: A review of serous tumors at our institution between 1995-2012 revealed 376 pure HGSC, 19 pure LGSC and 15 HGSC apparently co-existing with LGSC. We selected this group with mixed features of LGSC and HGSC, 15 cases with pure HGSC and 19 with pure LGSC using MD Anderson diagnosis criteria. Low pass whole genome sequencing (WGS) was performed using DNA from tumor areas enriched by macrodissection from FFPE tissue sections. In mixed cases, low and high grade areas were extracted separately. A dedicated CNV bioinformatics pipeline named Wandy processed the data. Coverage profiles were segmented into broad- and focal-CNV events to facilitate low- versus high-grade classifications, using a regression-tree approach.
Results: All pure LGSC cases harbor broad CNV events related to aneuploidy, with few to no focal amplifications or loss. In contrast, HGSC cases tend to have “spike-like” focal events on top of aneuploidy events. In the group with mixed features of HGSC and LGSC more heterogeneity was seen: 27% (4/15) showed progression from LGSC to HGSC and 73% showed either pure low grade (33%, 5/11) or pure high grade (40%, 6/11) genotype in both high grade and low grade components.
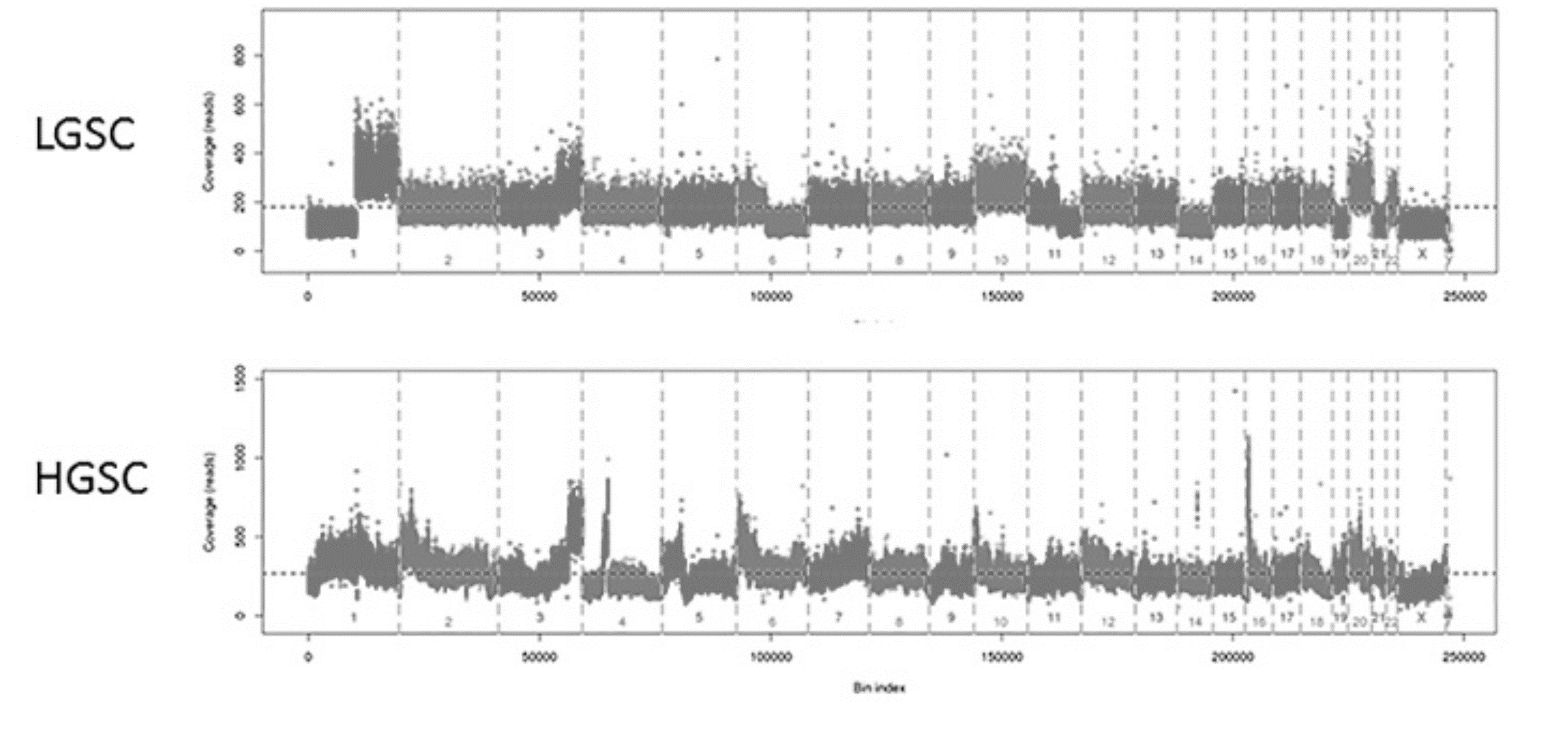
Conclusions: Low-pass WGS may be used as a tool to distinguish pure LGSC from pure HGSCs. We found that the total number of focal CNV events could be a single metric to clearly separate pure LGSC and HGSC cases. Mixed grade carcinomas showed more heterogeneity, suggesting only some of these tumors represent progressive low to high grade disease. Additional analysis is needed to determine if CNV metrics correlate with prognosis in morphologically ambiguous cases.
|