|
---------------------------- 2022 Platform Presentation Winner (shared) --------------------------
[768] Molecular Correlates of Invasion Pattern in HPV-Associated Endocervical Adenocarcinoma: Emergence of Two Distinct Risk-Stratified Tiers
Aarti Sharma, Anjelica Hodgson, Brooke Howitt, Bojana Djordjevic, Kay Park, Marisa Nucci, Carlos Parra-Herran
Background: The pattern-based (Silva) classification of invasive HPV-associated endocervical adenocarcinomas (HPVA) is an established and reproducible method to predict outcomes for this otherwise stage-dependent group of tumors. Previous studies utilizing targeted sequencing have shown a correlation between mutational profiles and invasive pattern. However, such correlation has not been explored using comprehensive molecular testing.
Design: Clinicopathologic data including invasive pattern (Silva groups A, B, and C) was collected for a cohort of invasive HPVA which underwent massive parallel sequencing using a panel covering 447 genes. Pathogenic alterations, molecular signatures, single-nucleotide polymorphisms, tumor mutational burden (TMB), and copy number alterations (CNA) were correlated with pattern of invasion.
Results: 45 HPVA (11 pattern A, 17 pattern B and 17 pattern C tumors, Table 1) were included. Patients with pattern A presented at stage I with no involved lymph nodes or evidence of recurrence (in those with > 2 months of follow-up). Patterns B and C patients also mostly presented at stage I with negative lymph nodes but had greater frequency of recurrence. 3/17 pattern B and 1/17 pattern C HPVAs harbored lymphovascular space invasion (LVI), respectively. APOBEC mutational signature was detected only in Silva pattern C tumors (5/17), and pathogenic PIK3CA alterations were detected only in destructively invasive HPVA (patterns B and C, Figure 1). When cases were grouped as low-risk (pattern A and pattern B without LVI) and high-risk (pattern B with LVI and pattern C), high-risk tumors were enriched in single-nucleotide variations in PIK3CA and ATRX, as well as ERBB2 amplification. There was a statistically significant difference in TMB between low-risk and high-risk pattern tumors (p=0.006), as well as between Pattern C tumors with and without an APOBEC signature (p=0.002). Volume of CNA increased along the progression from pattern A to C (Figure 2).
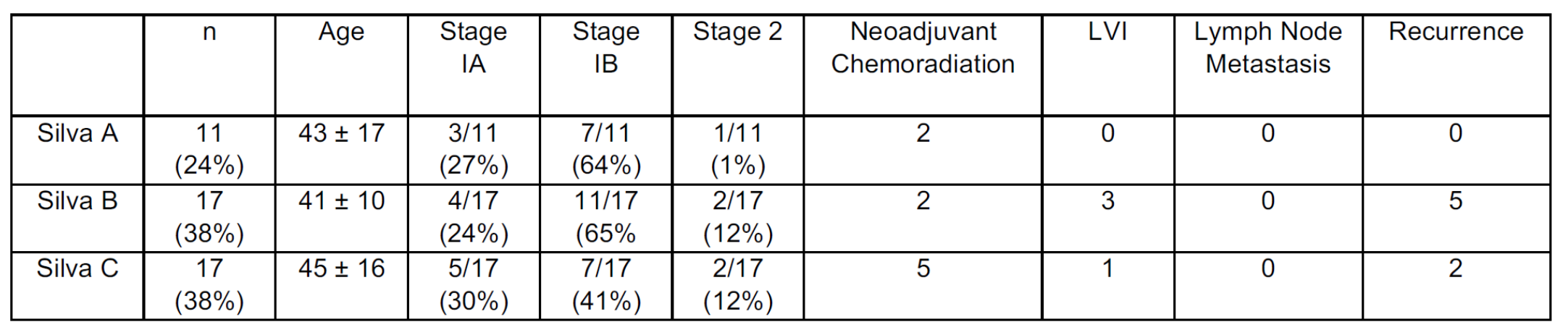
Conclusions: Our findings corroborate the hypothesis that key molecular events in HPVA correlate with the morphologic invasive properties of the tumor and their aggressiveness. Pattern B tumors with LVI clustered with pattern C tumors, whereas pattern B tumors without LVI approached pattern A genotypically. Our study provides a biologic basis for consolidating the Silva system into low-risk (pattern A + B without LVI) and high-risk (pattern B with LVI and pattern C) categories.
|